Here
is an example to use SEGID.
Start
SEGID from the webpage by clicking the button "start SEGID", a window will
pop up:
Now,
input a multiple sequence alignment to get it work. Click button 'Input',
then click button 'Input Alignment' in the pop-up dialog. Or, choose menu
'input'>'input Alignment'. Now you can an input dialog. Input the alignment
in the text area. You may
1. Type directly in the textarea, (Edit operations are the same as common
text editors.) OR
2. Copy & Paste from an existing file. (If you have a file containing
the alignment, load the file in any
text editor, and copy the content of file. Eg. open the file with Notepad,
choose menu 'edit'>
'select all', then choose menu 'edit'>'copy' to copy the alignment data
you want to input. Now, return
to SEGID and move focus to the textarea in input dialog by clicking in
the textarea. Press Ctrl+V
to paste alignment. Or, if under Unix, click the middle button of mouse.)
Choose 'protein' or 'DNA' according to your data, also choose the appropriate data format.
Finally, click 'submit' to submit the alignment.
The alignment format SEGID recognizes includes FASTA, CLUSTAL, GCG-MSF, and Stockholm. For each format, an example alignment is provided. It can be loaded into input textarea by clicking button 'load example' in the input dialog and then choosing corresponding format. For example, following is a multiple sequence alignment of Clustal format including 9 sequences.
CLUSTAL W (1.81) multiple sequence alignment CARP CCAGGACGACTAAATCAAGCCGCCTTTATTGCCTCACGCCCAGGGGTCTTTTACGGACAA LOACH CCAGGACGCCTTAACCAAACCGCCTTTATTGCCTCCCGCCCCGGGGTATTCTATGGGCAA CHICKEN CCTGGACGACTAAATCAAACCTCCTTCATCACCACTCGACCAGGAGTGTTTTACGGACAA COW CCAGGCCGTCTAAACCAAACAACCCTTATATCGTCCCGTCCAGGCTTATATTACGGTCAA WHALE CCAGGACGCCTAAACCAAACAACCTTAATATCAACACGACCAGGCCTATTTTATGGACAA SEAL CCAGGACGACTAAACCAAACAACCCTAATAACCATACGACCAGGACTGTACTACGGTCAA MOUSE CCAGGCCGACTAAATCAAGCAACAGTAACATCAAACCGACCAGGGTTATTCTATGGCCAA RAT CCCGGCCGCCTAAACCAAGCTACAGTCACATCAAACCGACCAGGTCTATTCTATGGCCAA HUMAN CCCGGACGTCTAAACCAAACCACTTTCACCGCTACACGACCGGGGGTATACTACGGTCAA ** ** ** ** ** *** * * * * * ** ** ** * * ** ** *** CARP TGCTCTGAAATTTGTGGAGCTAATCACAGCTTTATACCAATTGTAGTTGAAGCAGTACCT LOACH TGCTCAGAAATCTGTGGAGCAAACCACAGCTTTATACCCATCGTAGTAGAAGCGGTCCCA CHICKEN TGCTCAGAAATCTGCGGAGCTAACCACAGCTACATACCCATTGTAGTAGAGTCTACCCCC COW TGCTCAGAAATTTGCGGGTCAAACCACAGTTTCATACCCATTGTCCTTGAGTTAGTCCCA WHALE TGCTCAGAGATCTGCGGCTCAAACCACAGTTTCATACCAATTGTCCTAGAACTAGTACCC SEAL TGCTCAGAAATCTGTGGTTCAAACCACAGCTTCATACCTATTGTCCTCGAATTGGTCCCA MOUSE TGCTCTGAAATTTGTGGATCTAACCATAGCTTTATGCCCATTGTCCTAGAAATGGTTCCA RAT TGCTCTGAAATTTGCGGCTCAAATCACAGCTTCATACCCATTGTACTAGAAATAGTGCCT HUMAN TGCTCTGAAATCTGTGGAGCAAACCACAGTTTCATGCCCATCGTCCTAGAATTAATTCCC ***** ** ** ** ** * ** ** ** * ** ** ** ** * ** ** CARP CTCGAACACTTCGAAAAC---------------------TGATCCTCATTAATACTAGAA LOACH CTATCTCACTTCGAAAAC---------------------TGGTCCACCCTTATACTAAAA CHICKEN CTAAAACACTTTGAAGCC---------------------TGATCCTCACTA--------- COW CTAAAGTACTTTGAAAAA---------------------TGATCTGCGTCAATATTA--- WHALE CTAGAAGTCTTTGAAAAA---------------------TGATCTGTATCAATACTA--- SEAL CTATCCCACTTCGAGAAA---------------------TGATCTACCTCAATGCTT--- MOUSE CTAAAATATTTCGAAAAC---------------------TGATCTGCTTCAATAATT--- RAT CTAAAATATTTCGAAAAC---------------------TGATCAGCTTCTATAATT--- HUMAN CTAAAAATCTTTGAAATA---------------------GGGCCCGTATTTACCCTATAG ** ** ** * * CARP GACGCCTCGCTAGGAAGCTAA LOACH GACGCCTCACTAGGAAGCTAA CHICKEN ---------CTGTCATCTTAA COW ------------------TAA WHALE ------------------TAA SEAL ------------------TAA MOUSE ------------------TAA RAT ------------------TAA HUMAN --------------------- You can load it by clicking button 'load example' in the input dialog, and then choose 'Clustal' in pop-up dialog.

SEGID reads the alignment, calculates a score for every column with chosen scoring scheme (default scoring is SP-score and IDENTITY matrix. Users can specify scoring method and matrix via 'set scoring scheme'.) Then the alignment is displayed, and conserved segments (high score substrings) are identified. Three algorithms for identifying conserved segments are provided.
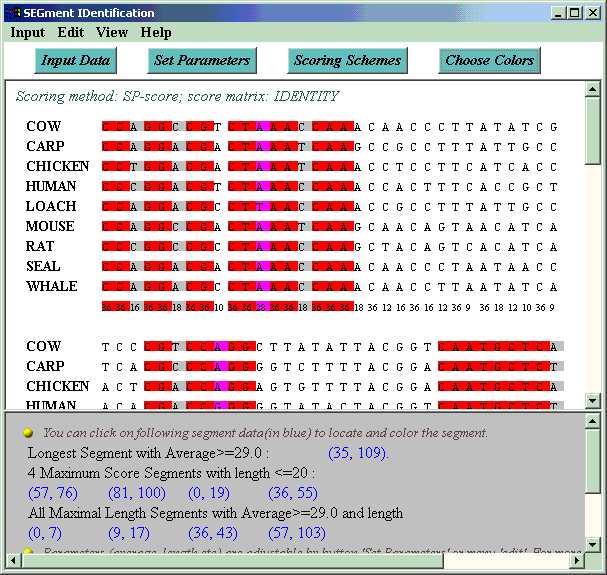
By default, all maximal length segments with average value and length lower bound are colored with pink in the alignment, among which columns of particularly poor scores (below the threshold set by user) are colored with lightgray, and good columns are colored with magenta. Users can also switch to other algorithms of interesting by clicking button 'choose algorithm' or choosing menu 'view'>'algorithm'. Accurate positions of all these segments are output at the bottom of the window. Users can locate a segment in the alignment by clicking on the segment position data.
For more information about how to use this software, please refer to "help" menu or button in SEGID.